
In cancer diagnosis or advanced treatments like immunotherapy, every detail in a medical image counts. Radiologists and pathologists rely on these images to track tumors, understand their boundaries, and analyze how they interact with surrounding cells. This work demands pinpoint accuracy across several tasks—identifying whether a tumor is present, locating it precisely, and mapping its contours on complex CT scans or pathology slides.
Yet, these crucial steps—object recognition, detection, and segmentation—are often tackled separately, which can limit the depth of analysis. Current tools like MedSAM (opens in new tab) and SAM (opens in new tab) focus on segmentation only, thus missing out on the opportunity to blend these insights holistically and relegating object as an afterthought.
In this blog, we introduce BiomedParse (opens in new tab), a new approach for holistic image analysis by treating object as the first-class citizen. By unifying object recognition, detection, and segmentation into a single framework, BioMedParse allows users to specify what they’re looking for through a simple, natural-language prompt. The result is a more cohesive, intelligent way of analyzing medical images that supports faster, more integrated clinical insights.
While biomedical segmentation datasets abound, there are relatively few prior works on object detection and recognition in biomedicine, let alone datasets covering all three tasks. To pretrain BiomedParse, we created the first such dataset by harnessing OpenAI’s GPT-4 for data synthesis from standard segmentation datasets (opens in new tab).
BiomedParse is a single foundation model that can accurately segment biomedical objects across nine modalities, as seen in Figure 1, outperforming prior best methods while requiring orders of magnitude fewer user operations, as it doesn’t require an object-specific bounding box. By learning semantic representation for individual object types, BiomedParse’s superiority is particularly pronounced in the most challenging cases with irregularly shaped objects. Through joint pretraining of object recognition, detection, and segmentation, BiomedParse opens new possibilities for holistic image analysis and image-based discovery in biomedicine.
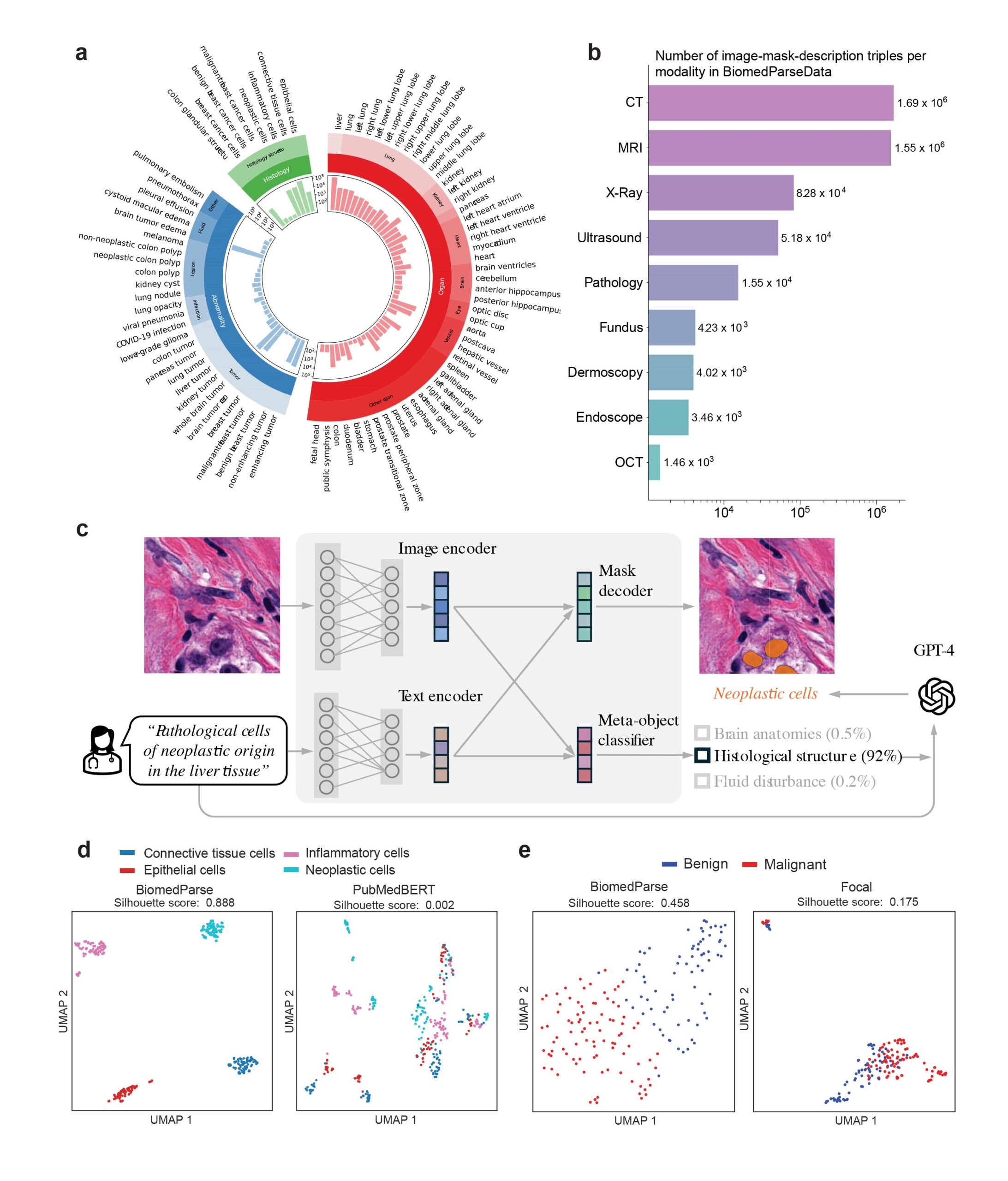
Image parsing: a unifying framework for holistic image analysis
Back in 2005, researchers first introduced the concept of “image parsing”—a unified approach to image analysis that jointly conducts object recognition, detection, and segmentation. Built on Bayesian networks, this early model offered a glimpse into a future of joint learning and reasoning in image analysis, though it was limited in scope and application. Fast forward to today, cutting-edge advances in generative AI have breathed new life into this vision. With our model, BiomedParse, we have created a foundation for biomedical image parsing that leverages interdependencies across the three subtasks, thus addressing key limitations in traditional methods. BiomedParse enables users to simply input a natural-language description of an object, which the model uses to predict both the object label and its segmentation mask, thus eliminating the need for a bounding box (Figure 1c). In other words, this joint learning approach lets users segment objects based on text alone.
Spotlight: AI-POWERED EXPERIENCE
Microsoft research copilot experience
Discover more about research at Microsoft through our AI-powered experience
Harnessing GPT-4 for large-scale data synthesis from existing datasets
We created the first dataset for biomedical imaging parsing by harnessing GPT-4 for large-scale data synthesis from 45 existing biomedical segmentation datasets (Figure 1a and 1b). The key insight is to leverage readily available natural-language descriptions already in these datasets and use GPT-4 to organize this often messy, unstructured text with established biomedical object taxonomies.
Specifically, we use GPT-4 to help create a unifying biomedical object taxonomy for image analysis and harmonize natural language descriptions from existing datasets with this taxonomy. We further leverage GPT-4 to synthesize additional variations of object descriptions to facilitate more robust text prompting.
This enables us to construct BiomedParseData, a biomedical image analysis dataset comprising over 6 million sets of images, segmentation masks, and text descriptions drawn from more than 1 million images. This dataset includes 64 major biomedical object types, 82 fine-grained subtypes, and spans nine imaging modalities.
State-of-the-art performance across 64 major object types in 9 modalities
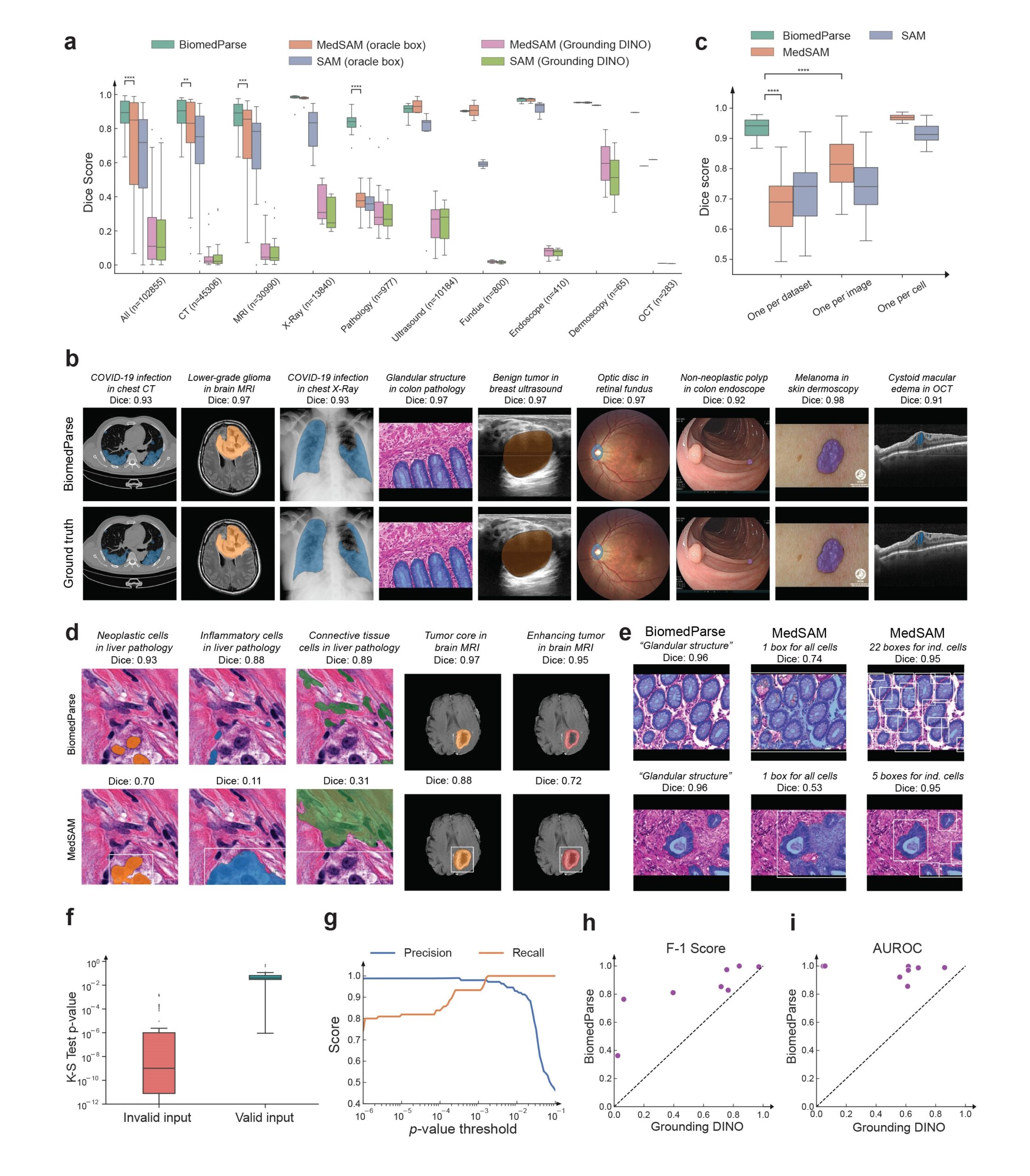
We evaluated BiomedParse on a large held-out test set with 102,855 image-mask-label sets across 64 major object types in nine modalities. BiomedParse outperformed prior best methods such as MedSAM and SAM, even when oracle per-object bounding boxes were provided. In the more realistic setting when MedSAM and SAM used a state-of-the-art object detector (Grounding DINO) to propose bounding boxes, BiomedParse outperformed them by a wide margin, between 75 and 85 absolute points in dice score (Figure 2a). BiomedParse also outperforms a variety of other prominent methods such as SegVol, Swin UNETR, nnU-Net, DeepLab V3+, and UniverSeg.
Recognizing and segmenting irregular and complex objects
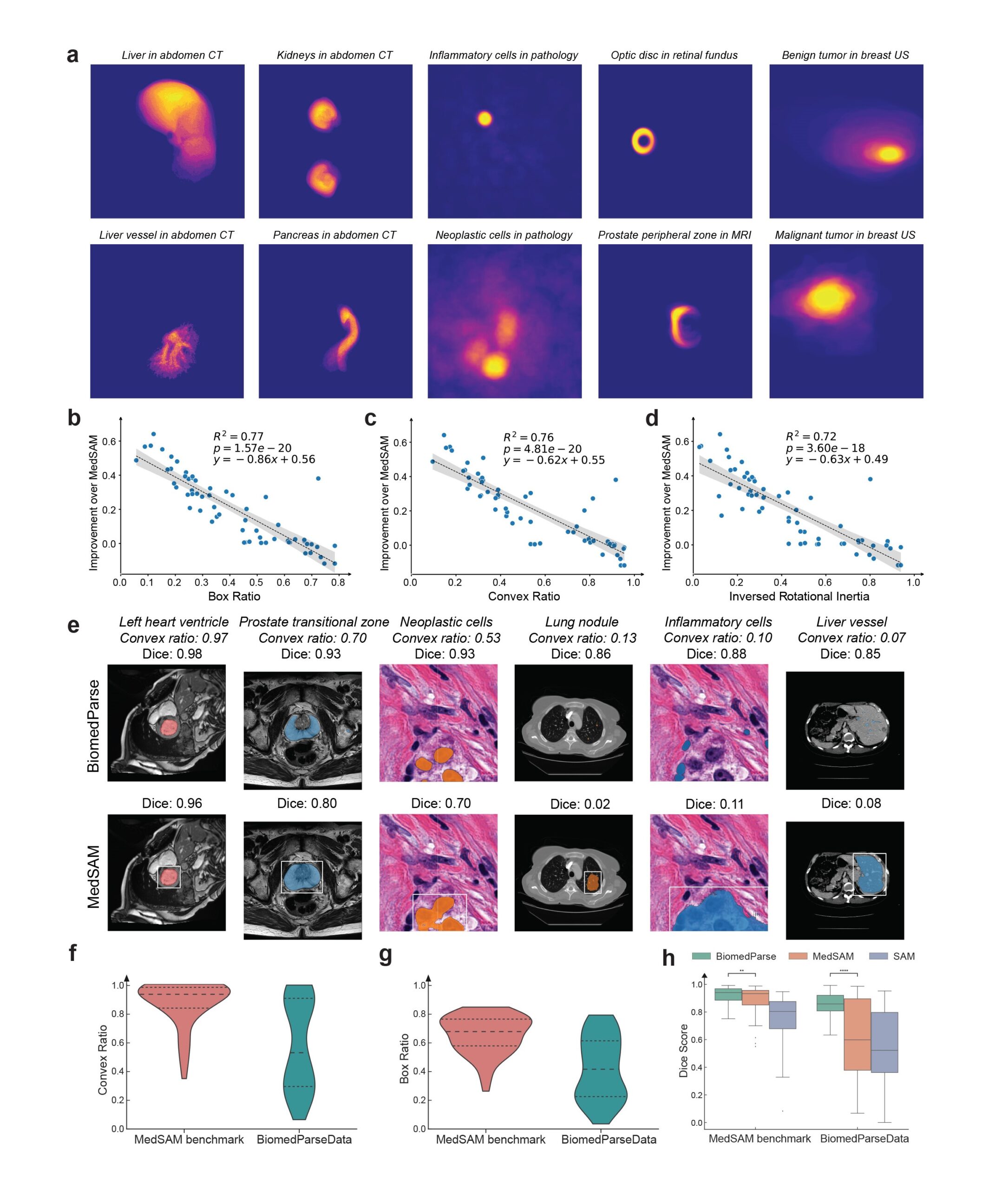
Biomedical objects often have complex and irregular shapes, which present significant challenges for segmentation, even with oracle bounding box. By joint learning with object recognition and detection, BiomedParse learns to model object-specific shapes, and its superiority is particularly pronounced for the most challenging cases (Figure 3). Encompassing a large collection of diverse object types in nine modalities, BiomedParseData also provides a much more realistic representation of object complexity in biomedicine.
Promising step toward scaling holistic biomedical image analysis
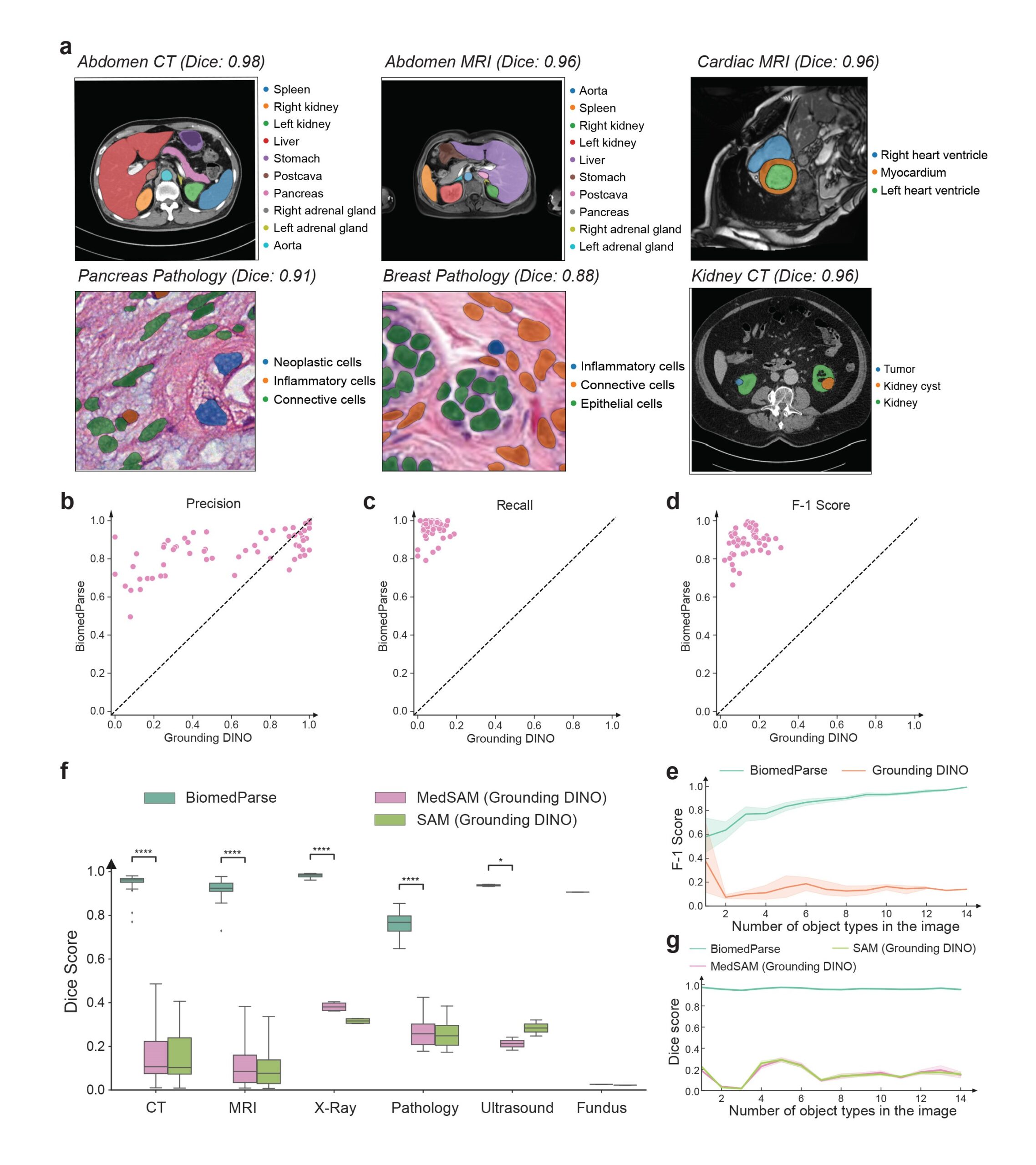
By operating through a simple text prompt, BiomedParse requires substantially less user effort than prior best methods that typically require object-specific bounding boxes, especially when an image contains a large number of objects (Figure 2c). By modeling object recognition threshold, BiomedParse can detect invalid prompt and reject segmentation requests when an object is absent from the image. BiomedParse can be used to recognize and segment all known objects in an image in one fell swoop (Figure 4). By scaling holistic image analysis, BiomedParse can potentially be applied to key precision health applications such as early detection, prognosis, treatment decision support, and progression monitoring.
Going forward, there are numerous growth opportunities. BiomedParse can be extended to handle more modalities and object types. It can be integrated into advanced multimodal frameworks such as LLaVA-Med (opens in new tab) to facilitate conversational image analysis by “talking to the data.” To facilitate research in biomedical image analysis, we have made BiomedParse open-source (opens in new tab) with Apache 2.0 license. We’ve also made it available on Azure AI (opens in new tab) for direct deployment and real-time inference. For more information, check out our demo. (opens in new tab)
BiomedParse is a joint work with Providence and the University of Washington’s Paul G. Allen School of Computer Science & Engineering, and brings collaboration from multiple teams within Microsoft*. It reflects Microsoft’s larger commitment to advancing multimodal generative AI for precision health, with other exciting progress such as GigaPath (opens in new tab), BiomedCLIP (opens in new tab), LLaVA-Rad (opens in new tab), BiomedJourney (opens in new tab), MAIRA (opens in new tab), Rad-DINO (opens in new tab), Virchow (opens in new tab).
(Acknowledgment footnote) *: Within Microsoft, it is a wonderful collaboration among Health Futures, MSR Deep Learning, and Nuance.
Paper co-authors: Theodore Zhao, Yu Gu, Jianwei Yang (opens in new tab), Naoto Usuyama (opens in new tab), Ho Hin Lee, Sid Kiblawi, Tristan Naumann (opens in new tab), Jianfeng Gao (opens in new tab), Angela Crabtree, Jacob Abel, Christine Moung-Wen, Brian Piening, Carlo Bifulco, Mu Wei, Hoifung Poon (opens in new tab), Sheng Wang (opens in new tab)
The post BiomedParse: A foundation model for smarter, all-in-one biomedical image analysis appeared first on Microsoft Research.
